Japanese Journal of Gastroenterology Research
Research Article - Open Access, Volume 2
Correlation between dysbiosis and incidence of gastrointestinal cancers
Sara AR1; Eslam MS1; Mohamed Ahmed Raslan1; Nagwa A Sabri2*
1Drug Research Centre, Cairo, Egypt.
2Department of Clinical Pharmacy, Faculty of Pharmacy- Ain Shams University, Cairo, Egypt.
*Corresponding Author : Nagwa Ali Sabri
Professor of Clinical Pharmacy Department, Faculty of Pharmacy, Ain Shams University, Cairo, Egypt.
Email: nagwa.sabri@yahoo.com &
nagwa.sabri@pharma.asu.edu.eg
Received : Jul 07, 2022
Accepted : Jul 29, 2022
Published : Aug 05, 2022
Archived : www.jjgastro.com
Copyright : © Sabri NA (2022).
Abstract
Background: The human gastrointestinal tract (GIT) population of almost 200 common bacteria, viruses, and fungi supplies the host with distinct metabolic activities and is critical in health and illness state.
Aim: The review goal is to correlate between dysbiosis and occurrence of different GIT diseases conditions. Furthermore, we will highlight on the effect of diet and probiotics on dysbiosis progression.
Method: Literature search was done and information was collected from trusted resources like PubMed and other journals.
Discussion: Microbiota have a wide range of effects on cancer risk, including host metabolism, immunological function, host/microbial sensing pathways, and cellular proliferation. Esophageal carcinoma is the most common cause of cancer mortality globally. On the other hand, gastric cancer is a complicated disease influenced by genetic, molecular, and environmental factors, with H. pylori infection being the most prevalent cause. The development of colorectal cancer (CRC) is highly challenging. Changes in gut microbiota patterns are consistently related with CRC, with tumor signatures diverging from adjacent normal tissue. The aetiology of inflammatory bowel disease (IBD) is known to be multifaceted, with an excessive immune response to gut flora in genetically susceptible individuals. Furthermore, hepatocellular carcinoma (HCC) is the most common primary liver cancer and the fourth largest cause of cancer deaths worldwide; both its incidence and mortality rate are rising. All the previously mentioned disease conditions are correlated with alterations in normal microbiota diversity and abundance. Diet habits and misuse of antibiotics definitely affect gut commensal ecosystem.
Conclusion: Dysbiosis is mainly correlated with different GIT disease conditions. Dietary habits have an influence of normal state of commensal gut microbiome. Probiotics showed to have a significant effect in enhancing gut ecosystem, and support management of different GIT disease conditions.
Keywords: GUT microbiota; Gastric cancer; Colorectal Cancer; Probiotics; Inflammatory Bowel Disease; Liver cancer; Lactobacillus acidophilus.
Abbreviations: GIT: Gastrointestinal Tract; DNA: Deoxyribonucleic Acid; ESCC: Esophageal Squamous Cell Carcinoma; EAC: Esophageal Adenocarcinoma; GERD: Gastroesophageal Reflux Disease; HFD: High-fat Diet; BE: Barrett Esophagus; G-CSF: Granulocyte Colony-Stimulating Factor; TLR: Toll-like Receptor; GC: Gastric Cancer; CRC: Colorectal Cancer; IBD: Inflammatory Bowel Diseases; CD: Including Mainly Crohn’s Disease; UC: Ulcerative Colitis; GOS: Galactooligosaccharides; HCC: Hepatocellular Carcinoma; TMA: Trimethylamine; SCFA: Short-chain Fatty Acids; SIBO: Small Intestine Bacterial Overgrowth; DCA: Deoxycholic Acid; LPS: Lipopolysaccharide; ALD: Alcoholic Liver Disease.
Citation: Sara AR, Eslam MS, Raslan MA, Sabri NA. Correlation between Dysbiosis and Incidence of GastrointestinalCancers. Japanese J Gastroenterol Res. 2022; 2(11): 1102.
Introduction
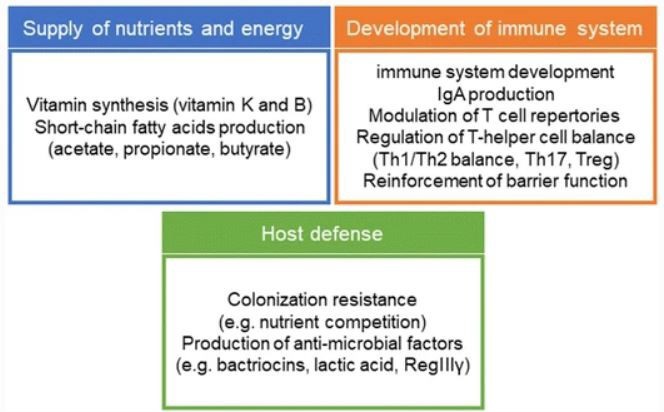
The term “microbiota” refers to the complete community of microorganisms that inhabit a certain region and includes not only bacteria, but also fungus, viruses, archaea, and protozoans [1]. The human gastrointestinal tract (GIT) population of nearly 200 common bacteria, viruses, and fungi provides unique metabolic activities to the host and is profoundly essential in health and diseases [2]. Furthermore, the gut microbiota helps in production of energy and nutrients (such as vitamins and short-chain fatty acids) that would otherwise be unavailable to the host and are required for mucosal barrier homeostasis [3]. The normal gut microbiota performs particular functions in host nutrition metabolism, xenobiotic and drug metabolism, gut mucosal barrier structural integrity, immune regulation, and pathogenic defence. Several variables influence shaping of gut flora. They include the method of delivery (vaginal or caesarean); nutrition during infancy (breast milk or formula feeds); and diet throughout maturity (vegan or meat based); and the usage of antibiotics or antibiotic-like substances originating from the environment or the gut commensal ecosystem [4]. There is a growing recognition of the gut microbiota’s role to tumor development, with certain infectious agents known to generate 15 to 20% of cancers, while other malignancies are connected to the collective gut microbiota with/without the presence of specific trigger organisms [3]. COVID-19 infection can change gut microbiota, causing dysbiosis, and increase gut permeability, leading to the advancement of secondary bacterial infections and bacterial pneumonia. Furthermore, dysbiosis has been linked to the development of inflammatory bowel disease, cardiovascular illness, and autoimmune disease. Dietary adjustments and supplements can help to improve symbiosis [5].
Materials and methods
Data search to investigate the association between gut microbiota dysbiosis and different gastrointestinal tract (GIT) cancer and diseases types. Furthermore, the effect of diet on microbiota diversity and GIT ecobalance is highlighted.
Discussion
Cancers of the Gastrointestinal tract
The impact of microbiota on cancer risk is multifaceted, including host metabolism, immunological function, host/microbial sensing pathways, and cellular proliferation [7]. Carbohydrate structures on gastrointestinal mucins, for example, serve as binding sites and/or metabolic substrates for bacteria and are essential factors in site-specific microbial colonization [8]. Increases in pro-inflammatory cytokine levels caused by microbes can cause epithelial DNA damage, including epigenetic regulatory alterations, resulting in genetic instability. These variables have an impact on cancer development, promotion, and spread, as well as therapy [3].
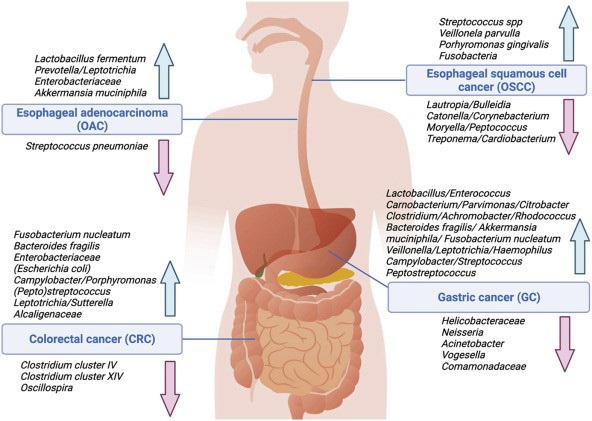
Esophageal Cancer is the leading cause of cancer death worldwide, having two different histologic forms which are, esophageal squamous cell carcinoma (ESCC), and esophageal adenocarcinoma (EAC) [9]. Globally, ESCC prevalence is declining, however EAC cases are rapidly increasing, owing to broad adoption of a Western diet and a rise in obesity, both of which contribute to gastroesophageal reflux disease (GERD), the primary risk factor for EAC. Barrett esophagus (BE), a precancerous lesion, usually precedes EAC [10]. When compared to normal esophageal tissue, ESCC, EAC, and BE have lower species diversity/richness. Campylobacter, Streptococcus, Prevotella, Veillonella, Leptotrichia, and Actinobacillus members with different microbial profiles linked with ESCC and EAC are more consistently abundant in BE. Streptococcus species, Veillonella parvula, and Porphyromonas gingivalis are the most prevalent in ESCC, while Lautropia, Bulleidia, Catonella, Corynebacterium, Moryella, Peptococcus, Treponema, and Cardiobacterium are few [11].
Diet also influences microbiota composition in esophageal carcinogenesis. A study indicated that the esophageal microbiota of Sprague Dawley rats on a high-fat diet (HFD) differs from that of rats fed a regular chow diet, with a rise in Clostridium species and a decrease in Escherichia, Shigella, and Lactobacillus genera [12]. Furthermore, it was shown that transgenic interleukin (IL)2–IL1β mice fed a high-fat diet (HFD) developed esophageal cancers faster than control mice. This acceleration was linked to changes in the gut microbiota as well as immunological modifications such as enhanced Toll-like receptor (TLR) expression, an increased ratio of neutrophils to natural killer cells, and abnormal levels of T-cell recruitment factors like chemokin (C-C motif) ligands 6 and 12 (CCL6, CCL12), granulocyte colony-stimulating factor (G-CSF), and chemokine (C-X-C motif) ligand 1 (CXCL1), resulting in an increase in immune cells that express the C-X-C motif chemokine receptor 2 (CXCR2) [13].
Surgical changes to the upper gastrointestinal tract result in functional changes that influence host metabolism and mucosal homeostasis. Esophagojejunostomy to induce BE in rats’ changes both TLR expression and the makeup of the esophageal microbiota, with a substantial drop in Lactobacillus and a rise in Clostridium, Enterococcus, and Streptococcus [14]. Furthermore, the addition of antibiotics exacerbates this impact, which is reversed by the addition of rebamipide, a mucosal protective drug used to treat peptic ulcer disease [15]. Riboflavin is another component that might influence the equilibrium between esophageal mucosal integrity and gut flora. In rats, riboflavin deficiency is associated with esophageal epithelial atrophy and decreased activity of xenobiotic metabolic pathways. Riboflavin supplementation in rats has a direct effect on gut microbiota composition, with a decrease in Firmicutes abundance and an increase in Proteobacteria [16].
Gastric Cancer (GC) is a complex illness with genetic, molecular, and environmental factors all impacting disease progression, with H. pylori infection being the most common cause [17]. Because of the acidic environment and other local antimicrobial characteristics, it was long assumed that the stomach was solely inhabited by H pylori and was unsuitable to other microbes [18]. Gastric carcinogenesis, however, is H pylori-independent beyond a certain point in the evolution of mucosal changes since colonization levels decline in individuals with intestinal metaplasia and dysplasia, and is practically absent by the adenocarcinoma stage. H. pylori may thus work in a hit-and-run fashion, preparing the stomach mucosa for future oncogenic alterations caused by other bacteria [19]. Helicobacter and, to a much lesser extent, Streptococcus, Prevotella, and Neisseria dominate microbiota profiles in patients with H pylori-induced superficial gastritis or even glandular atrophy, resulting in decreased phylotype richness, diversity, and evenness compared to patients with normal gastric mucosa [20]. As a result of the loss of specialized glandular tissue and reduced acid production in GC tissue, H pylori is eliminated and intestinal commensals such as Lactobacillus, Enterococci, Carnobacterium, Parvimonas, Citrobacter, Clostridium, Achromobacter, and Rhodococcus are enriched. [21] as well as oral species; Fusobacterium nucleatum, Veillonella, Leptotrichia, Haemophilus, and Campylobacter. [22] Furthermore, species, including F nucleatum, are associated with worse prognosis [23].
Colorectal Cancer (CRC) development is extremely difficult. CRC is consistently associated with changes in gut microbiota profiles, with tumor signatures deviating from neighboring normal tissue. Differences include decreased variety and changed community structure, which become more pronounced as the CRC advances [24]. Lower numbers of beneficial, potentially protective taxa, such as butyrate-producing species from Clostridium clusters IV and XIV, have been repeatedly documented in CRC, whereas an increase in species such as Fusobacterium, Bacteroides, Campylobacter, Escherichia, and Porphyromonas has been linked to increased pro-oncogenic capacity [25]. Firmicutes, Actinobacteria phyla and the Lachnospiraceae family are more commonly found in premalignant adenomas, while Proteobacteria, Alcaligenaceae, Entero-bacteriaceae, and Sutterella species are more common in CRC [26]. F nucleatum is commonly found in CRC tissue, both at the adenoma and adenocarcinoma stages, along with other oral commensal species such as Peptostreptococcus, Leptotrichia, and Campylobacter species [27]. Enterotoxigenic Bacteroides fragilis (ETBF) and Escherichia coli (which are found to promote colon carcinogenesis in colitis-associated cancer rather than sporadic CRC), Streptococcus gallolyticus subspecies gallolyticus, and Enterococcus faecalis have also been linked in CRC pathogenesis. The presence of B fragilis/ETBF in CRC tissue also is likewise related with a worse prognosis [28].
Inflammatory Bowel Disease and Crohn's Disease
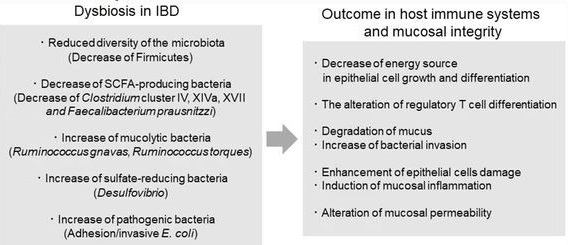
Inflammatory bowel diseases (IBD), which mostly include Crohn's disease (CD) and ulcerative colitis (UC), are a category of chronic inflammatory gastrointestinal disorders [29]. Inflammatory bowel diseases (IBD) pathophysiology is known to be complicated, with an overactive immune response to gut microbiota in genetically predisposed people. Patients with IBD show lower biodiversity and relative abundance of butyrate-producing bacterial genera such as Faecalibacterium, Bifidobacterium, and Lactobacillus [30]. Furthermore, several studies have shown that the shape of mucosa-associated bacterial communities in the gut differs from that seen in faeces, and changes in microbial diversity have been observed between inflammatory and non-inflammatory areas of the gut [31]. F. prausnitzii, a Clostridium cluster IV member, has been shown to produce butyrate, which has an anti-inflammatory effect. When compared to healthy persons, F. prausnitzii, Blautia faecis, Roseburia inulinivorans, Ruminococcus torques, and Clostridium lavalense levels are lower in CD patients [6].
Clinical studies revealed that the genera Enterococcus and Hydrogenophilus were more abundant in the faecal and mucosal samples of patients with ulcerative colitis (UC) and Crohn's disease (CD) than in healthy controls, whereas Janibacter, Lactobacillus, and Schlegelella were enriched in the mucosal samples of UC or CD patients, respectively. There were 16 and 234 differently abundant genera in faecal and biopsy samples from UC patients, respectively, and 31 and 344 differentially abundant genera in faecal and biopsy communities from CD patients, respectively [31]. Consequently, the mucosal communities had much more differentially abundant characteristics than the faecal microbiota. Studies discovered that 50 and 63 Proteobacteria genera were enriched in the mucosal microbiota communities of patients with ulcerative colitis (UC) and Crohn's disease (CD), respectively, showing an expansion of Proteobacteria in the gut of IBD patients, particularly during an active phase of the disease [32]. The makeup of the gut flora is also affected by IBD medication. When compared to untreated individuals, mesalazine lowers faecal bacteria and the concentration of mucosal adherent bacteria [33]. Furthermore, it suppresses the development of Mycobacterium avium subspecies paratuberculosis, which has been connected to the pathogenesis of CD [34]. Another study found that mesalazine reduces gene expression in Salmonella enterica serovar Typhimurium that is related with bacterial invasiveness and antibiotic resistance, perhaps promoting the onset of IBD following infection [35]. Patients with inflammatory bowel disease (IBD) have a considerably higher risk of colorectal cancer (CRC), owing to chronic intestinal inflammation's pro-neoplastic effects [36]. Colorectal cancer (CRC) is a leading cause of death in both ulcerative colitis (UC) and colonic Crohn's disease (CD), contributing for 10 to 15% of all-cause mortality in inflammatory bowel disease patients (IBD) [37].
The age at IBD diagnosis was a key risk factor for CRC compared with background population, as those diagnosed at age 0 to 19 had a RR of 43.8 (95% CI 27.2 to 70.7) compared with those diagnosed at age 20 to 39 with a RR of 2.65 (95% CI 1.97 to 3.56); both were referenced against those diagnosed older than 40 years [38]. In advanced disease stages or in cases that are in need for intensive care unit, itopride (a prokinetic drug) is well tolerated and effective in critically sick individuals with Enteral feeding intolerance (EFI) which is a frequent problem. Enteral feeding intolerance (EFI) is associated with poor clinical outcomes leading to worse prognosis in terms of mortality [39].
Liver cancer: Hepatocellular carcinoma (HCC) is the most prevalent primary liver cancer and the fourth leading cause of cancer-related deaths globally; both its incidence and mortality rate are increasing. HCC has a dismal prognosis, and treatment choices are restricted. The liver is physically and physiologically linked to the gastrointestinal system, which is our body's greatest reservoir of microorganisms. A growing body of data suggests that communication between the liver and the gut (and its microbiome) plays a role in the development of chronic liver disease to liver cancer [40]. The liver receives the majority of its blood supply from the GI tract via the portal circulation, which is rich in important nutrients that support the metabolic activity of the liver in the Gut-Liver Axis. In turn, the intestine gets several metabolites from the liver, such as bile acids, which are generated in the liver and transferred into the gut, where they influence the composition of the intestinal microbiome owing to their antibacterial characteristics [41]. The intestinal barrier is a multilayered defence mechanism that protects the body from foreign infections. Finally, the gut-vascular barrier prevents intestinal bacteria from spreading into the systemic circulation. Despite this, food antigens and bacterial components that have survived the intestinal mucosal/vascular barrier and entered the liver can be found in the enterohepatic circulation. As a result, the liver serves as the body's second firewall, relying on its powerful immune system to identify and clear gut-derived toxins (including bacteria), safeguarding the host and maintaining whole-body homeostasis [42]. The gut microbiota promotes host health by being metabolically and immunologically active. This involves nutrition extraction from indigestible fibres, tissue homeostasis preservation, and pathogen prevention. This link is reciprocal; nutrition, lifestyle, age, antibiotics, and disease progression may all influence the microbiota [43]. The liver influences the makeup of the gut microbiome by synthesizing and transporting bile acids (which have antimicrobial characteristics) into the intestine, where IgA antibodies are produced. The intestinal microbiota metabolizes liver-derived bile acids into secondary products, which are then recycled into the liver [44]. Following meal fermentation, the gut microbiota produces a range of metabolites, including trimethylamine (TMA), vitamins, and short-chain fatty acids (SCFA), which regulate host immunity, barrier function, and liver function [45].
During chronic illness, small intestine bacterial overgrowth (SIBO) and changes in microbiome composition (i.e., dysbiosis) occurs, which is linked with increased intestinal permeability (leaky gut) permit endotoxins into the systemic circulation, which corresponds with disease development [40].
The abundance of Enterobacteriaceae and Proteobacteria was higher in colonic biopsies from alcoholic liver disease (ALD) patients than in biopsies from healthy individuals, but Bacteroidetes and Firmicutes were lower; In ALD patients, the presence of potentially harmful bacteria such as Enterobacteriaceae, Veillonelaceae, and Streptococcus was higher, but Lachnospiraceae was lower. Patients' microbiome makeup varied according to illness severity; levels of Bifidobacteria and Streptococci were greatest in severe alcohol hepatitis cases, but Enterobacteria was enriched in all instances [40]. When compared to nonalcoholic steatohepatitis (NASH)-cirrhotic patients, microbiome study revealed that hepatocellular carcinoma (HCC) patients from the NASH aetiology had a different makeup. The hepatocellular carcinoma (HCC) microbiome was associated with lower diversity, significantly higher abundances of anti-inflammatory bacterial genera (e.g., Bifidobacterium and Blautia), and significantly higher abundances of the genera Enterococcus, Ruminococcus, Bacteroides, Phascolarctobacterium, and Oscillospira [46].
Furthermore, clinical studies indicated that the microbiome composition of patients with HCC progressing from viral hepatitis differed from that of nonrelated viral hepatitis-HCC patients and controls; viral hepatitis-HCC patients had a higher species richness and were enriched in Prevotella when compared to both nonrelated viral hepatitis-HCC patients and healthy controls. When compared to viral hepatitis-HCC patients, nonrelated viral hepatitis-HCC patients were dominated by proinflammatory bacterial genera such as Enterococcus and Escherichia-Shigella, with reduced abundances of Ruminococcus and Faecalibacterium [47]. Activation of the Toll-Like Receptor (TLR4) signalling pathway in response to Lipopolysaccharide (LPS), as well as the direct toxic effects of secondary bile acids (microbial metabolites) in the liver, have been found in clinical studies to increase HCC development [48]. Secondary bile acids have also been demonstrated to influence immunological function and the development of HCC. Deoxycholic acid (DCA), for example, led to liver inflammation by boosting the senescence-associated secretory phenotype (SASP), ultimately leading to the development of obesity-related HCC. A further investigation found that DCA activated mTOR, which caused nonalcoholic steatohepatitis (NASH)-associated HCC [40]. Changes in the bile acid pool following antibiotic therapy (a decrease in secondary bile acid and an increase in primary bile acid) resulted in improved antitumor immunity (NKT cell activation after primary bile acid production; TbMCA). Antibiotics also dramatically lowered DCA levels and, as a result, liver inflammation and the development of NASH-related HCC [40].
Hepatocellular carcinoma (HCC) may develop or recur in chronic hepatitis C (CHC) genotype 4 cirrhotic individuals on sofosbuvir/daclatasvir and ribavirin. Liver cirrhosis was showed to be the cause of HCC development [49].
Table 1: Summary of Microbiome dysbiosis and disease clinical outcomes.
Disease |
Microbiota dysbiosis |
Clinical Outcome |
Esophageal Cancer |
Most prevalent Depleted |
-Barrett esophagus (BE). -Esophageal squamous cell carcinoma (ESCC). -Esophageal adenocarcinoma (EAC). |
Gastric Cancer |
Abundant Depleted |
-Loss of specialized glandular tissue and reduced acid production. -Gastric carcinoma. |
Colorectal Cancer |
Abundant Depleted |
-Increased pro-oncogenic capacity. -Colorectal carcinoma. |
Inflammatory Bowel Disease and Crohn's Disease |
Abundant
Depleted |
-Induction of mucosal inflammation. -Alteration of mucosal permeability. -Crohn's Disease. -Ulcerative colitis. |
Liver Cancer |
Abundant Depleted |
-Increased intestinal permeability (leaky gut) permit endotoxins into the systemic circulation - Hepatocellular carcinoma (HCC) |
Dietary effect on gut microbiota
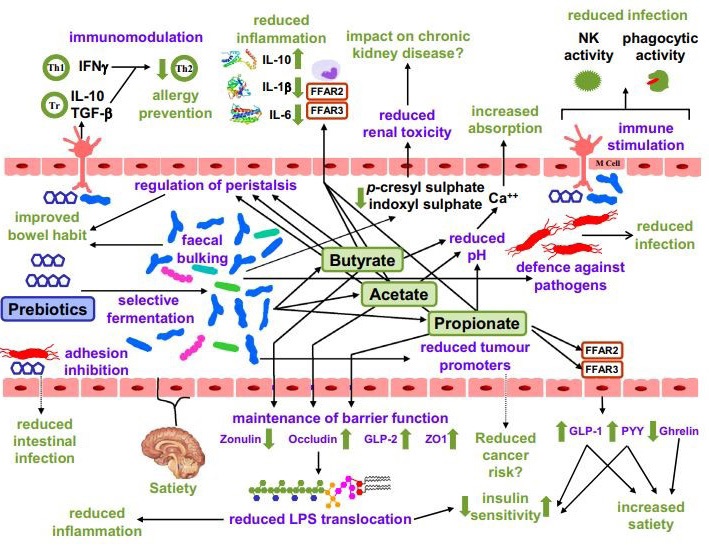
Dietary prebiotics have been defined as “a selectively fermented ingredient that results in specific changes in the composition and/or activity of the gastrointestinal microbiota, thus conferring benefit(s) upon host health” [50]. Fermentation of dietary prebiotics in the gut involves metabolic cross-feeding where the products of fermentation by one or more bacterial species provide the substrate(s) for other bacterial species. This complex cooperative activity of the gut microbiota is essential for good health. Bacterial fermentation of amino acids and proteins, which occurs mainly in the distal colon, generates a range of metabolites, many of which have a toxic potential. These include hydrogen sulphide, branched-chain fatty acids (BCFAs), phenol, indole, p-cresol, indoxylsulfate, p-cresylsulfate, and ammonia [51]. Several studies have demonstrated modulation of colonic microbiota by prebiotic inulin or inulin-type fructans. Real-time polymerase chain reaction (PCR) identification of selected bacterial species in the feces of human volunteers after inulin ingestion showed that the prevalence of Faecalibacterium prausnitzii and two Bifidobacterium species, B. adolescentis and B. bifidum, increased significantly [52].
Lactation causes substantial alterations in the gut microbiota, followed by a subsequent shift with the introduction of solid meals. Until roughly 2-3 years of age, the newborn is vulnerable to limited bacterial diversity and a high rate of microbial flux. This time is essential for the formation of the gut microbiota, with abnormalities associated to an increased risk of autoimmune disorders and metabolic problems later in life [53]. Dietary preferences are acknowledged as a significant environmental element influencing the host's microbial community makeup. Long-term dietary habits have a greater impact on Firmicutes, Bacteroides, and Prevotella ratios in the gut than short-term alterations [54]. A rigorous vegan or vegetarian diet, on the other hand, is related with a significant drop in the number of Enterobacteriaceae, Bacteroides, and Bifidobacterium species [55]. The capacity of the gut microbiota to adjust quickly to dietary changes may be representative of our volatile hunter-gatherer nutritional intake, which was predicated on the requirement for dietary flexibility with periods of feast and famine [56]. One longitudinal study involving daily gut microbiota investigations of two individuals over the course of a year found that changes in fibre intake are positively correlated with a change in abundance of 15% of the microbial community the following day [57]. These relatively rapid changes to the gut microbiota could be a ‘shock reaction’ to an influx of incoming nutrients, possibly causing a transient disruption of microbial composition. The ability of the gut microbiota to cope with this stress is part of the inherent plastic nature of the normal microbiota. In this way, the gut microbiota is able to adapt and adopt a new beneficial or detrimental state when faced with a continuous perturbation [53]. Children fed with a high-fiber, plant-based diet in Burkina Faso had a markedly divergent gut microbial composition compared with their European counterparts, whose daily diet was richer in sugar, fat, and protein [58].
In addition, a diet rich in red meat aggravated dextran sodium sulfate (DSS)-induced colitis in mice, resulting in higher disease activity and histopathological scores. Therefore, alterations in the gut microbiota community composition and function as a result of westernized diet may trigger and maintain autoimmunity by promoting the emergence of pathogens [31]. Among the 13 enriched genera in the UC mucosal microbiota, Clostridium XI had the greatest number of correlations with diet, being correlated to 10 dietary nutrients, while among the 13 genera with decreased abundance, Dorea had the most correlations, with 13 dietary nutrients. In the CD group Schlegelella was positively correlated with nine dietary nutrients, while Coprococcus had the greatest number of negative correlations, with associations observed for a further nine dietary nutrients. In the fecal microbiota 23 genera (17 increased and six decreased) in UC and 18 genera (7 increased and 11 decreased) in CD samples were significantly correlated with diet [31].
The genera Acinetobacter, Bacteriovorax, Bdellovibrio, and Brevundimonas were significantly correlated with diet and were enriched in the UC and CD groups. In patients with CD Acinetobacter was negatively correlated with protein, vitamin B1, phosphorus, and copper. Bacteriovorax was positively related to calcium, while Bdellovibrio was positively related to vitamin D [31]. Among the dietary groups, fish and shellfish consumption had the greatest number of correlations with mucosa-associated microbiota in patients with IBD. In addition, cereal and fruits were correlated with most genera in the fecal microbiota of patients with IBD. With respect to dietary nutrients, the intake of vitamin A and total energy had the greatest number of correlations with fecal and mucosal microbiota communities, respectively, in patients with UC [31]. In faeces, vitamin A was linked to Chryseobacterium, Odoribacter, Phenylobacterium, and Rhizobium. In mucosal microbiota communities, energy intake was linked to Blautia, Clostridium XI, Dorea, Fusicatenibacter, Odoribacter, Roseburia, and Schlegelella. Vitamin D consumption had the most associations with both the mucosal and faecal microbiota in CD patients, and it was also favorably connected to the abundance of Fusicatenibacter in both mucosal and faecal samples. Overall, gut microbiota was clearly connected to food and nutrient consumption in IBD patients [31].
A 2018 systematic review and meta-analysis of 64 research examined the influence of fibre on gut flora. Dietary fibre strategies, notably fructans and galacto-oligosaccharides (GOS), were observed to enhance the number of Bifidobacterium and Lactobacillus species in the faeces but had no effect on alpha-diversity [59]. It was discovered by Liu et al. that eating fructo-oligosaccharide (FOS) and GOS for 14 days enhanced Bifidobacterium while decreasing butyrate-producing bacteria in 35 healthy people. However, after a 28-day washout phase, the gut microbiota was demonstrated to revert to its pre-intervention baseline condition, demonstrating that the observed microbial modifications are lost within the 28-day washout period if these prebiotic fibres are not consumed [60]. A clinical study showed that nicotinamide at 1000 mg daily was tolerated, decreased metabolic anomalies, and increased quality of life in diabetic nonalcoholic fatty liver disease patients while having no effect on liver fibrosis or steatosis [61]. Burton et al., also found that there was an absence of the probiotic bacterial strains related to the two-week consumption of probiotic yoghurt after a three week wash-out period (n = 14) [62]. In a study by Kellingray et al., increased consumption of Brassica was associated with reduced relative abundance of sulphate-producing bacteria and members of Rikenellaceae, Ruminococcaceae, Mogibacteriaceae, and Clostridiales [63]. Findings have indicated the benefit of probiotics in aiding the treatment of infectious- and antibiotic-associated diarrhea, insulin resistance in diabetes, and remission and maintenance of inflammatory bowel disease, amongst others [53].
Probiotics have been defined as “live microorganisms which, when administered in adequate amounts, confer a health benefit on the host”. Many probiotic bacteria are traditionally used in the fermentation of food but are now predominantly ingested by the public as supplement-like probiotic products that contain live bacteria. After consumption probiotics have the capacity to colonize and proliferate within the gastrointestinal tract thereby influencing the gut ecosystem [53]. Plant-based foods such as fruit, vegetables, legumes, grains and nuts contain dietary fibre. While fibre as a whole is generally accepted to be beneficial to gastrointestinal health, specific dietary fibre types including inulin, FOS and GOS are also considered to be prebiotic; defined as “a substrate that is selectively used by host microorganisms conferring a health benefit” [64]. Extensive research has shown that probiotics have anti-proliferative or pro-apoptotic actions in GI malignancies, with colonic cancer cells and gastric cancer cells being the most widely examined [65]. Previous research found that L. rhamnosus GG strain suppressed the growth of both human gastric cancer cells and colonic cancer cells, whereas Bifidobacterium adolescentis SPM0212 decreased the proliferation of three human colon cancer cell lines, including HT-29, SW 480, and Caco-2. Bacillus polyfermenticus, L. acidophilus 606, LGG/Bb12, and LGG/Bifidobacterium animalis subsp. lactis were among the other probiotic products or strains that shown anticancer activity against human colon cancer cells [66]. Few randomized controlled trials have been conducted to investigate the effect of probiotics on the prevention and inhibition of intestinal carcinogenesis [67]. Probiotics' advantages extend beyond the prevention of intestinal malignancies to the prevention of symptoms and problems in cancer patients having colorectal surgery and receiving intestinal cancer therapy [68].
Table 2: Probiotic microorganisms used in human nutrition [66].
Type Lactobacillus |
Type Bifidobacterium |
Lactic Acid Bacteria |
Other Microorganisms |
L. acidophilus (a) |
B. adolescentis (a) |
Enterococcus faecium (a) |
Bacillus clausii (a) |
L. amylovorus (b) |
B. animalis (a) |
Lactococcus lactis (b) |
Escherichia coli Nissle 1917(a) |
L. casei (a), (b) |
B. bifidum (a) |
Streptococcus thermophiles (a) |
Saccharomyces cerevisiae (boulardi) (a) |
L. gasseri (a) |
B. breve (b) |
||
L. helveticus (a) |
B. infantis (a) |
||
L. johnsonii (b) |
B. longum (a) |
||
L. pentosus (b) |
|||
L. plantarum (b) |
|||
L. reuteri (a) |
|||
L. rhamnosus (a), (b) |
Conclusion
Dysbiosis is correlated to the incidence of different types of gastrointestinal tract cancer and disease conditions. Dietary habits have an influence of normal state of commensal gut microbiome. Probiotics and dietary supplements have a significant effect in enhancing gut microbiome diversity and abundance. Besides, supporting in management of different gastrointestinal tract disease conditions.
Declarations
Conflicts of interest: The authors have no conflicts of interest to declare.
Funding sources: This review did not receive any fund.
Acknowledgments: The author would like to thank Drug Research Center for its support in data collection and manuscript writing.
References
- Sekirov I, Russell SL, Antunes LC, Finlay BB. Gut microbiota in health and disease. Physiol Rev. 2010; 90: 859904.
- Valdes AM, Walter J, Segal E, Spector TD. Role of the gut microbiota in nutrition and health. BMJ 2018; 361: k2179.
- Smet A, Kupcinskas J, Link A, Hold GL, Bornschein J. The Role of Microbiota in Gastrointestinal Cancer and Cancer Treatment: Chance or Curse? Cell Mol Gastroenterol Hepatol. 2022; 13: 857-874.
- Jandhyala, Sai Manasa et al. “Role of the normal gut microbiota.” World journal of gastroenterology vol. 21,29 (2015): 8787-803.
- Nagwa A Sabri, et al. “Microbiota in the Era of COVID-19. Correlation and Benefits”. Acta Scientific Pharmaceutical Sciences 6.7 (2022): 22-29.
- Nishida A, Inoue R, Inatomi O, et al. Gut microbiota in the pathogenesis of inflammatory bowel disease. Clin J Gastroenterol. 2018; 11: 1-10.
- Louis P, Hold GL, Flint HJ. The gut microbiota, bacterial metabolites and colorectal cancer. Nat Rev Microbiol. 2014; 12: 661-672
- Silva E, Teixeira A, David L, Carneiro F, Reis CA, Sobrinho-Simões J, et al. Mucins as key molecules for the classification of intestinal metaplasia of the stomach. Virchows Arch. 2002; 440: 311-317
- Arnold M, Soerjomataram I, Ferlay J, Forman D. Global incidence of oesophageal cancer by histological subtype in 2012. Gut. 2015; 64: 381-387
- Gregson EM, Bornschein J, Fitzgerald RC. Genetic progression of Barrett’s oesophagus to oesophageal adenocarcinoma. Br J Cancer. 2016; 115: 403-410
- Liu Y, Lin Z, Lin Y, Chen Y, Peng XE, He F, Liu S, et al. Streptococcus and Prevotella are associated with the prognosis of oesophageal squamous cell carcinoma. J Med Microbiol. 2018; 67: 1058-1068
- Kaakoush NO, Lecomte V, Maloney CA, Morris MJ. Cross-talk among metabolic parameters, esophageal microbiota, and host gene expression following chronic exposure to an obesogenic diet. Sci Rep 2017; 7: 45753.
- Ferreira RM, Pereira-Marques J, Pinto-Ribeiro I, Costa JL, Carneiro F, Machado JC, Figueiredo C. Gastric microbial community profiling reveals a dysbiotic cancer-associated microbiota. Gut 2018; 67: 226–236.
- Liu X, Shao L, Liu X, Ji F, Mei Y, Cheng Y, et al. Alterations of gastric mucosal microbiota across different stomach microhabitats in a cohort of 276 patients with gastric cancer. EBioMedicine 2019; 40: 336–348.
- Hu Y-L, Pang W, Huang Y, Zhang Y, Zhang C-J. The gastric microbiome is perturbed in advanced gastric adenocarcinoma identified through shotgun metagenomics. Front Cell Infect Microbiol 2018; 8: 433.
- Rajilic-Stojanovic M, Figueiredo C, Smet A, Hansen R, Kupcinskas J, et al. Systematic review: gastric microbiota in health and disease. Aliment Pharmacol Ther 2020; 51: 582-602.
- Hansen ABR, Johannesen TB, Spiegelhauer MR, et al. Distinct composition and distribution of the gastric mycobiota observed between dyspeptic and gastric cancer patients evaluated from gastric biopsies. Microb Heal Dis 2020; 2: e340
- Gao JJ, Zhang Y, Gerhard M, Mejias-Luque R, Zhang L, et al. Association between gut microbiota and Helicobacter pylori-related gastric lesions in a high-risk population of gastric cancer. Front Cell Infect Microbiol 2018; 8: 202.
- Yang I, Woltemate S, Piazuelo MB, Bravo LE, Yepez MC, Romero-Gallo J, Delgado AG, Wilson KT, Peek RM, Correa P, Josenhans C, Fox JG, Suerbaum S. Different gastric microbiota compositions in two human populations with high and low gastric cancer risk in Colombia. Sci Rep 2016; 6: 18594.
- Arita S, Inagaki-Ohara K. High-fat-diet-induced modulations of leptin signaling and gastric microbiota drive precancerous lesions in the stomach. Nutrition 2019; 67-68: 110556
- Nakatsu G, Li X, Zhou H, Sheng J, Wong SH, Wu WKK, Ng SC, Tsoi H, Dong Y, Zhang N, He Y, Kang Q, Cao L, Wang K, Zhang J, Liang Q, Yu J, Sung JJY. Gut mucosal microbiome across stages of colorectal carcinogenesis. Nat Commun 2015; 6: 8727.
- Feng Q, Liang S, Jia H, Stadlmayr A, Tang L, Lan Z, Zhang D, Xia H, Xu X, Jie Z, Su L, Li X, Li X, Li J, Xiao L, Huber-Schönauer U, Niederseer D, Xu X, Al-Aama JY, Yang H, Wang J, Kristiansen K, Arumugam M, Tilg H, Datz C, Wang J. Gut microbiome development along the colorectal adenoma-carcinoma sequence. Nat Commun 2015; 6: 6528
- Drewes JL, White JR, Dejea CM, Fathi P, Iyadorai T, Vadivelu J, Roslani AC, Wick EC, Mongodin EF, Loke MF, Thulasi K, Gan HM, Goh KL, Chong HY, Kumar S, Wanyiri JW, Sears CL. High-resolution bacterial 16S rRNA gene profile meta-analysis and biofilm status reveal common colorectal cancer consortia. NPJ Biofilms Microbiomes 2017; 3: 34.
- Mori G, Rampelli S, Orena BS, Rengucci C, De Maio G, Barbieri G, Passardi A, Casadei Gardini A, Frassineti GL, Gaiarsa S, Albertini AM, Ranzani GN, Calistri D, Pasca MR. Shifts of faecal microbiota during sporadic colorectal carcinogenesis. Sci Rep 2018; 8: 10329.
- Yu T, Guo F, Yu Y, Sun T, Ma D, Han J, Qian Y, Kryczek I, Sun D, Nagarsheth N, Chen Y, Chen H, Hong J, Zou W, Fang J-Y. Fusobacterium nucleatum promotes chemoresistance to colorectal cancer by modulating autophagy. Cell 2017; 170:548-563.e16.
- Boleij A, Hechenbleikner EM, Goodwin AC, Badani R, Stein EM, Lazarev MG, Ellis B, Carroll KC, Albesiano E, Wick EC, Platz EA, Pardoll DM, Sears CL. The Bacteroides fragilis toxin gene is prevalent in the colon mucosa of colorectal cancer patients. Clin Infect Dis 2015; 60: 208-215.
- Purcell RV, Pearson J, Aitchison A, Dixon L, Frizelle FA, Keenan JI. Colonization with enterotoxigenic Bacteroides fragilis is associated with early-stage colorectal neoplasia. PLoS One 2017; 12: e0171602.
- Buc E, Dubois D, Sauvanet P, Raisch J, Delmas J, Darfeuille-Michaud A, Pezet D, Bonnet R. High prevalence of mucosa-associated E. coli producing cyclomodulin and genotoxin in colon cancer. PLoS One 2013; 8: e56964.
- Ng SC. Emerging leadership lecture: inflammatory bowel disease in Asia: emergence of a “Western” disease. J Gastroenterol Hepatol. 2015; 30: 440-445.
- Wright EK, Kamm MA, Teo SM, Inouye M, Wagner J, Kirkwood CD. Recent advances in characterizing the gastrointestinal microbiome in Crohn’s disease: a systematic review. Inflamm Bowel Dis. 2015; 21: 1219-1228.
- Weng YJ, Gan HY, Li X, Huang Y, Li ZC, Deng HM, Chen SZ, Zhou Y, Wang LS, Han YP, Tan YF, Song YJ, Du ZM, Liu YY, Wang Y, Qin N, Bai Y, Yang RF, Bi YJ, Zhi FC. Correlation of diet, microbiota and metabolite networks in inflammatory bowel disease. J Dig Dis. 2019 Sep; 20: 447-459.
- Rooks MG, Veiga P, Wardwell-Scott LH, et al. Gut microbiome composition and function in experimental colitis during active disease and treatment-induced remission. ISME J. 2014; 8: 1403-1417.
- Andrews CN, Griffiths TA, Kaufman J, et al. Mesalazine (5-aminosalicylic acid) alters faecal bacterial profiles, but not mucosal proteolytic activity in diarrhoea-predominant irritable bowel syndrome. Aliment Pharmacol Ther. 2011; 34: 374-83.
- Naser SA, Sagramsingh SR, Naser AS, et al. Mycobacterium avium subspecies paratuberculosis causes Crohn’s disease in some inflammatory bowel disease patients. World J Gastroenterol. 2014; 20: 7403-15.
- Schultz BM, Paduro CA, Salazar GA, et al. A potential role of salmonella infection in the onset of inflammatory bowel diseases. Front Immunol. 2017; 8: 191.
- Stidham RW, Higgins PDR. Colorectal Cancer in Inflammatory Bowel Disease. Clin Colon Rectal Surg. 2018 May; 31: 168-178.
- Wanders LK, Dekker E, Pullens B, Bassett P, Travis SPL, East JE. Cancer risk after resection of polypoid dysplasia in patients with longstanding ulcerative colitis: a meta-analysis. Clin Gastroenterol Hepatol 2014; 12: 756-764.
- Jess T, Simonsen J, Jørgensen KT, Pedersen BV, Nielsen NM, Frisch M. Decreasing risk of colorectal cancer in patients with inflammatory bowel disease over 30 years. Gastroenterology 2012; 143: 375-81.
- Elmokadem EM, El Borolossy RM, Bassiouny AM, Hanna MG, Darweesh EAG, Sabri NA. The efficacy and safety of itopride in feeding intolerance of critically ill patients receiving enteral nutrition: a randomized, double-blind study. BMC Gastroenterol. 2021 Mar 19; 21: 126.
- Moreno-Gonzalez M, Beraza N. The Role of the Microbiome in Liver Cancer. Cancers, Basel. 2021 May 12;13(10):2330.
- Grüner, N, Mattner, J. Bile Acids and Microbiota: Multifaceted and Versatile Regulators of the Liver–Gut Axis. Int. J. Mol. Sci. 2021; 22: 1397.
- Balmer, M.L, Slack, E, De Gottardi, A, Lawson, M.A.E, Hapfelmeier, S, Miele, L, Grieco, A, Van Vlierberghe, H, Fahrner, R, Patuto, N, et al. The Liver May Act as a Firewall Mediating Mutualism Between the Host and Its Gut Commensal Microbiota. Sci. Transl. Med. 2014; 6: 237ra66.
- Sommer, F, Bäckhed, F. The Gut Microbiota–Masters of Host Development and Physiology. Nat. Rev. Microbiol. 2013; 11: 227–238.
- Wahlström, A, Sayin, S.I, Marschall, H.-U, Bäckhed, F. Intestinal Crosstalk between Bile Acids and Microbiota and Its Impact on Host Metabolism. Cell Metab. 2016; 24: 41–50.
- Blacher, E, Levy, M, Tatirovsky, E, Elinav, E. Microbiome-Modulated Metabolites at the Interface of Host Immunity. J. Immunol. 2017: 198: 572–580.
- Ponziani FR, Bhoori, S, Castelli, C, Putignani, L, Rivoltini L, et al. Hepatocellular Carcinoma Is Associated with Gut Microbiota Profile and Inflammation in Nonalcoholic Fatty Liver Disease. Hepatology 2019; 69: 107–120.
- Liu, Q, Li, F, Zhuang, Y, Xu, J, Wang, J, et al. Alteration in Gut Microbiota Associated with Hepatitis B and Non-hepatitis Virus Related Hepatocellular Carcinoma. Gut Pathog. 2019: 11: 1–13.
- Seki, E, De Minicis, S, Osterreicher, C.H, Kluwe, J, Osawa, Y, et al. TLR4 Enhances TGF-beta Signaling and Hepatic Fibrosis. Nat. Med. 2007; 13: 1324–1332.
- Essawy, Aya & Mehrez, Mai & Shaheen, Sara & Garem, Hassan & et al. (2022). New incidence or recurrence hepatocellular carcinoma (HCC) in genotype 4 hepatitis C virus treated with sofosbuvir/daclatasvir with or without ribavirin. F1000Research. 10. 1105. 10.12688/f1000 research.73076.2.
- Gibson GR, Scott KP, Rastall RA, Tuohy KM, Hotchkiss A, et al. Dietary prebiotics: current status and new definition. Food Sci Technol Bull Funct Foods. 2011; 7: 1–19.
- Mohajeri, M Hasan et al. The role of the microbiome for human health: from basic science to clinical applications. European journal of nutrition vol. 2018; 57: 1-14.
- Ramirez-Farias C, Slezak K, Fuller Z, Duncan A, Holtrop G, Louis P. Effect of inulin on the human gut microbiota: stimulation of Bifidobacterium adolescentis and Faecalibacterium prausnitzii. Br J Nutr. 2009; 101: 541–550
- Leeming, E.R, Johnson, A.J, Spector, T.D, Le Roy, C.I. Effect of Diet on the Gut Microbiota: Rethinking Intervention Duration. Nutrients. 2019; 11: 2862.
- Wu GD, Chen J, Hoffmann C, et al. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011; 334: 105-108.
- Zimmer J, Lange B, Frick JS, et al. A vegan or vegetarian diet substantially alters the human colonic faecal microbiota. Eur J Clin Nutr. 2012; 66: 53-60.
- David LA, Maurice CF, Carmody RN, Gootenberg, DB, Button JE, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature 2014, 505, 559–563.
- David LA, Materna AC, Friedman J, Campos-Baptista MI, et al. Host lifestyle affects human microbiota on daily timescales. Genome Biol. 2014; 15: R89.
- De Filippo C, Cavalieri D, Di Paola M, et al. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc Natl Acad Sci U S A. 2010; 107: 14691-14696.
- So D, Whelan K, Rossi M, Morrison M, Holtmann G, Kelly JT, Set al. Dietary fiber intervention on gut microbiota composition in healthy adults: A systematic review and meta-analysis. Am. J. Clin. Nutr. 2018; 107: 965–983.
- Liu F, Li P, Chen M, Luo Y, Prabhakar M, et al. Fructooligosaccharide (FOS) and Galactooligosaccharide (GOS) Increase Bifidobacterium but Reduce Butyrate Producing Bacteria with Adverse Glycemic Metabolism in healthy young population. Sci. Rep. 2017; 7: 11789.
- El-Kady RR, Ali AK, El Wakeel LM, Sabri NA, Shawki MA. Nicotinamide supplementation in diabetic nonalcoholic fatty liver disease patients: randomized controlled trial. Therapeutic Advances in Chronic Disease. 2022.
- Burton KJ, Rosikiewicz M, Pimentel G, Butikofer U, von Ah U, et al. Probiotic yogurt and acidified milk similarly reduce postprandial inflammation and both alter the gut microbiota of healthy, young men. Br. J. Nutr. 2017; 117: 1312–1322
- Kellingray L, Tapp HS, Saha S, Doleman JF, Narbad A, et al. Consumption of a diet rich in Brassica vegetables is associated with a reduced abundance of sulphate-reducing bacteria: A randomised crossover study. Mol. Nutr. Food Res. 2017.
- Ferrarese, R, Ceresola, E, Preti, A, Canducci, F. Probiotics, prebiotics and synbiotics for weight loss and metabolic syndrome in the microbiome era. Eur. Rev. Med. Pharmacol. Sci. 2018; 22: 7588–7605.
- Liong MT. Roles of probiotics and prebiotics in colon cancer prevention: Postulated mechanisms and in-vivo evidence. Int J Mol Sci. 2008; 9: 854–63
- Javanmard A, Ashtari S, Sabet B, Davoodi SH, Rostami-Nejad M, Esmaeil Akbari M, Niaz A, Mortazavian AM. Probiotics and their role in gastrointestinal cancers prevention and treatment; an overview. Gastroenterol Hepatol Bed Bench. 2018; 11: 284-295.