Japanese Journal of Gastroenterology Research
Research Article - Open Access, Volume 2
Association between visfatin serum level and rs9939609 as a common polymorphism of fat mass and obesity: Associated gene in obese type 2 diabetic women
Davoud Jafari-Gharabaghlou1; Nosratollah Zarghami2*
1Department of Clinical Biochemistry and Laboratory Medicine, Faculty of Medicine, Tabriz University of Medical Sciences, Tabriz, Iran.
2Department of Medical Biochemistry, Faculty of Medicine, Istanbul Aydin University, Istanbul, Turkey.
*Corresponding Author : Nosratollah Zarghami
Professor of Medical Biochemistry, Faculty of
Medicine, Istanbul Aydin University, Istanbul, Turkey.
Email: nsoltanahmadi@aydin.edu.tr
Received : Mar 26, 2022
Accepted : Apr 29, 2022
Published : May 04, 2022
Archived : www.jjgastro.com
Copyright : © Zarghami N (2022).
Abstract
Background: Obesity is a well-understood health problem worldwide and is one of the main risk factors of health complications. The fat mass and obesity-associated (FTO) gene, located on chromosome 16q12.2 and might act as an important regulator of energy homeostasis, body weight, and hypothetically relates to increased risk of obesity by means of affecting the regulation of food intake. In this study, the association between visfatin level and FTO Polymorphism (rs9939609) in diabetic obese women in comparison to non-diabetic obese women was evaluated.
Methods: After collecting blood samples from 39 diabetics and 37 non-diabetic obese women. The age of women, body mass index (BMI), systolic blood pressure, diastolic blood pressure, fasting blood sugar, insulin, insulin resistance, low-density lipoprotein cholesterol, high-density lipoprotein cholesterol, glycated hemoglobin (HbA1c), triglycerides (TG), and visfatin level (using enzyme-linked immunosorbent assay) were measured. The FTO rs9939609 was amplified with specific primers and PCR amplicons were sequenced and three genotypes (AA, AT, and TT) were determined.
Results: There was a significant difference between visfatin level, FBS, insulin level, HOMA, and HbA1c of the two groups (P<0.05). The frequency of rs9939609 was high in the diabetic group. Adjusted analysis with rs9939609 showed the mean of TC, HbA1c and TG were significantly different among three genotypes (TT, AT, and AA). In diabetic group, significant moderate positive correlation was found between visfatin level and FBS (r=0.447, p= 0.004), TG (r = 0.390, p = 0.014) and SBP (r =-0.520, p = 0.001). The frequency of rs3399609 was high in diabetic obese women. In addition, the mean visfatin level in diabetic women who harbored rs3399609 was higher than wild-type genotype (TT).
Conclusions: Finally, it is concluded that visfatin could be used as a significant indicator in diabetic obese women.
Keywords: Visfatin; Obesity; Single nucleotide polymorphism; Diabetes mellitus type 2; rs3399609.
Abbreviations: D: Diabetic Women; ND: Non-Diabetic Women; FBS: Fast Blood Sugar; HbA1c: Glycated Hemoglobin; TG: Triglycerides; TC: Total Cholesterol; HDL: HighDensity Lipoprotein. LDL; Low-Density Lipoprotein; BMI: Body Mass Index; SBP: Systolic Blood Pressure; DBP: Diastolic Blood Pressure; WHR: Waist / Hip Ratio.
Citation: Jafari-Gharabaghlou D, Zarghami N. Association between visfatin serum level and rs9939609 as a common polymorphism of fat mass and obesity: Associated gene in obese type 2 diabetic women. Japanese J Gastroenterol Res. 2022; 2(7): 1079.
Introduction
Obesity is a well-understood health problem worldwide and is one of the main risk factors of several health complications like type 2 diabetes mellitus (T2DM), hypertension, metabolic syndrome, cardiovascular disorders, and also cancer [1-4]. The fat mass and obesity associated (FTO) gene, located on chromosome 16q12.2 encoding a 2-oxoglutarate-dependent nucleic acid demethylase, is mainly expressed in hypothalamus, pituitary, and adrenal glands [5] and might act as an important regulator of energy homeostasis [6], body weight [7,8] and hypothetically relates to increased risk of obesity by means of affecting the regulation of food intake [9]. For the first time, three genome-wide association studies were performed simultaneously and identified that FTO is a candidate locus that had a close association with obesity [8,10,11]. According to the studies, rs9939609 SNP located in the first intron of FTO gene is in association with body fat mass regulation through lipolysis [12]. Several studies showed the relationship between FTO SNPs and expanded risk of obesity by 1.2 - 1.3 fold in Europeans [13] and by 1.25 fold in Asians [14]. Its A allele seems to be related to raised risk of obesity, BMI, T2DM [8,15] among European populations [8,11,16-19], while for Asians this relevance is currently controversial in different studies and remains unclear [20-23]. Therefore, more studies among Asian populations seem substantial in order to address this challenge. Also, T2DM is a major international complication which initially was considered to exist in western countries but nowadays occurs in almost every nation worldwide and Asia alone accounts for nearly 60% of the diabetic population in the world nevertheless [23]. According to the previous studies, it would be acceptable that obesity have a strong association with metabolic disorders which diabetes was considered as a main of them [23]. Although further alterations for BMI obliterated the association of FTO gene with T2DM, it was primarily suggested as a type 2 diabetes-susceptibility gene. Thus, FTO was speculated to be mainly an obesity-susceptibility locus [24]. Several studies have shown the significant risk of T2DM associated with FTO locus even after alterations for BMI [25-30] whereas others failed to approve this [22,31-34].
Moreover, surplus amount of adipose tissue, which as an active endocrine organ secretes certain adipocytokines like leptin, adiponectin, resistin, TNFα, and IL-6, is known as a major factor in the pathogenesis of T2DM [35-40]. Visfatin, one of those adipocytokines, is a highly expressed cytokine in visceral fat which is known as pre-B cell colony-enhancing factor [41,42] and also nicotin amide phosphoribosyl-transferase [43]. Previous studies have postulated that it has a role in innate immunity, progression of colorectal cancer, proliferation of gastric cancer cell line (AGS ) and enhance telomerase reverse transcriptase gene expression [44-47]. A meta-analysis study conducted by Chen et al in 2006 strongly revealed increased plasma visfatin level (PVL) in obese and T2DM patients [44]. Considering controversial viewpoints toward visfatin and FTO role in T2DM patients in various populations, this study aims at revealing the association of rs9939609 SNP in FTO gene with serum level of visfatin in Iranian obese women suffering from T2DM.
Methods and materials
This study was performed from 2015-2016 during a year in The Immam Reza Teaching Hospital, Faculty of Medicine, Tabriz University of Medical Sciences, Tabriz, Iran.
Subjects and samples
Sample size of the study was calculated using G power version 3.0.10 (power: 80% and α: 0.05). In this case-control study, a total of 76 obese women (39 diabetic obese women and 37 non-diabetic obese women) were selected. The inclusion criteria for participations were: BMI > 30 Kg/m2 , FBS > 126 mg/ dL, FBS>126 mg/dL, age at the range of 30-60 years, no specific disease or chronic history, and no special diet at least in past 6 months. The patients who had kidney disease and users of lipid-lowering drugs were excluded from the study. Nondiabetic subjects were identified by having a fasting plasma glucose level lower than 110 mg/dL while T2DM subjects were diagnosed considering WHO criteria [48]. This study is approved by ethics committee of Tabriz University of Medical sciences, Tabriz, Iran (No: 19864/5). Informed consent was obtained from each subject before involving in the study. Two ml of peripheral blood samples were obtained in EDTA tubes and centrifuged at 5000 ×g for 10 min to separate the plasma and mononuclear cells. After separation, the samples were stored at -20OC until further steps. In this study serum was considered for biochemical analysis.
DNA extraction and rs9939609 SNP of FTO gene analysis
Total genomic DNA of white blood cells was extracted using saturated salting out protocol [49]. DNA pellets were dissolved in 50 μl of DNAse free double distilled water, checked for quality using NonDrop spectrophotometer, and then stored in -80oC until next steps. Using Oligo 7 software specific primer set (forward: 5´GTAGGAATACTAGGAGAGGAG3´and reverse: 5´GCTTAAAGTTAATGGCTTCAGG3´) were designed [50]. Polymerase chain reactions (PCR) were conducted in 25μl volumes as follows: 40ng of genomic DNA, 0.4μmol/L of each primer and 12.5μl of 1X master mix (CinnaGen Co., Iran), and then the reactions were adjusted to 25μl with deionized distilled water. PCR program was as follows: initial denaturation (95°C for 2 min) then 30 cycles of 95OC for 30 sec, 60OC for 30sec and 72OC for 40 sec and the final extension (72OC for 5 min). After checking the acceptable quality of PCR products by agarose gel electrophoresis, they were subjected to DNA sequencing (Lab Tech, Germany). Subsequent analysis included multiple sequence alignment using the BLAST program [51] (available at: https://blast.ncbi. nlm.nih.gov/Blast.cgi) and Chormas software (version 2.6.2).
Biochemical parameters and serum insulin and visfatin analysis
All samples and solutions were let to reach room temperature one hour before starting the assay. Age, both systolic and diastolic blood pressure (SBP, DBP), BMI, and FBS (fasting blood sugar) were calculated for each subject. Using ELISA technique the insulin and visfatin levels were assessed in the serum of subjects according to the standard protocol for ELISA assay. NycoCard ELISA (Switzerland HbA1c, U-Albumin CRP, D-Dimer) reader was used to conduct the assay. Total cholesterol (TC), high density lipoprotein (HDL) and triglyceride (TG) levels were measured using specific kits (ParsAzmun, Tehran, Iran) according to the available protocol. Low density lipoprotein (LDL) was calculated using Friedewlad equation [52] . Glycated hemoglobin (HbA1c), insulin, and visfatin serum levels were determined using NycoCard reader and enzyme-linked immunosorbent assay (ELISA: Crystal day Biotech Co., Ltd Kit). HOMA IR (insulin resistance) was calculated as Fasting Insulin × Fasting glucose /405. The body fat percentage was measured based on the WHR (Waist / hip ratio) and following formula (by BMI, age, and gender (male=1, female=0)) [53].
Adult Body Fat % = (1.20 × BMI) + (0.23 ×Age) – (10.8 × gender) – 5.4.
Statistical analysis
Data was analyzed using SPSS v.16 (SPSS Inc., Chicago, IL, USA). The student T-test was used to analyze the differences of assessed parameters between groups. The Pearson correlation coefficient test was used to analyze any correlation between clinico-pathological findings and gene polymorphism. P value <0.05 was assumed statistically significant.
Results
Characteristics of study population
The demographic, metabolic, and biochemical characteristics of these two groups are shown in (Table 1). There were statistically significant differences (p < 0.05) between diabetic (D) and non-diabetic (ND) groups considering age, FBS, insulin, HOMA and HbA1c values. The differences between mean level of visfatin in serum of diabetic and non-diabetic subjects were statistically significant (P =0.001) where its level was much higher in non-diabetics.
Table 1: Demographic, biochemical and metabolic characteristics of diabetic and non-diabetic groups.
Parameter |
(D), n=39 |
(ND) n=37 |
P value |
Age (year) |
54.64±9.68 |
47.38±10.63 |
0.003* |
FBS (mg/dL) |
177.64±48.03 |
87.94±9.53 |
0.001* |
HbA1c (%) |
7.86±1.71 |
4.71±0.36 |
0.001* |
TG (mg/dL) |
194.74±69.82 |
177.44±62.43 |
0.268 |
TC (mg/dL) |
190.54±40.01 |
202.69±44.12 |
0.432 |
HDL (mg/dL) |
48.95±10.48 |
47.25±11.47 |
0.591 |
LDL (mg/dL) |
149.72±61.18 |
150.14±50.51 |
0.928 |
BMI (kg/m2) |
33.39±8.04 |
32.56±3.06 |
0.695 |
Visfatin(ng/L) |
95.25 ± 30.87 |
169.25 ± 30.87 |
0.001* |
SBP (mmHg) |
119.73±1.64 |
116.21±25.58 |
0.406 |
DBP (mmHg) |
80.27±1.64 |
74.87±17.64 |
0.061 |
Insulin (μIU/mL) † |
21.40±4.48 |
24.46±12.50 |
0.004* |
HOMA † |
9.31±1.90 |
5.05±2.53 |
0.001* |
Fat mass (%) |
57.83±5.39 |
51.51±3.55 |
0.251 |
WHR |
1.08±0.39 |
1.40±0.65 |
0.014* |
rs9939609SNP genotyping
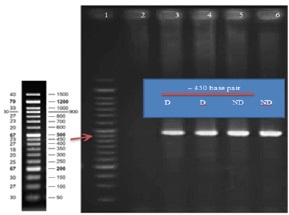
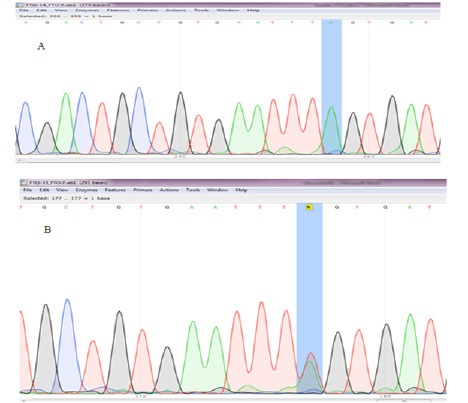
Figure 1 shows the agarose gel electrophoresis for amplified regions (450bp) and Figure 2 indicates Sanger sequencing. Table 2 demonstrates the frequency of alleles and genotypes. The A allele was more frequent among cases while the T allele was the most frequent allele of control subjects. Odds ratio was 90% and indicating that the odds of rs9939609 SNP occurrence in diabetic obese women is 90 times more than its occurrence in non-diabetic obese women.
Table 2: rs9939609 allele and genotype frequency in diabetic and non-diabetic obese women.
Group |
Genotype (No.) |
Frequency (%) |
Allele (No.) |
Frequency (%) |
Diabetics |
TT (3) |
7.7 |
T=31 |
T=39.74 |
TA (25) |
64.1 |
- |
- |
|
AA (11) |
28.2 |
A=47 |
A=60.24 |
|
Non-diabetics |
TT (30) |
81.1 |
T=66 |
T=89.19 |
TA (6) |
16.2 |
- |
- |
|
AA (1) |
2.7 |
A=8 |
A=10.81 |
The association of rs9939609 polymorphism with analyzed characteristics of subjects is shown in Table 3. Significant different were detected between three genotypes and TC, HbA1cvalue (in D group) and visfatin level and TG (in ND group).
Table 3: Association between rs993960 polymorphism and analyzed parameters.
Parameter |
(D) |
(ND) |
||||||
TT(3) |
TA(25) |
AA(11) |
P value |
TT(30) |
TA(6) |
AA(1) |
P value |
|
Age (year) |
67.00 ± 9.53 |
54.04 ± 8.77 |
52.64 ± 10.11 |
0.061 |
74.79 ± 10.84 |
46.83 ± 11.87 |
47.00 |
0.980 |
FBS (mg/dL) |
120.67 ± 16.01 |
183.28 ± 46.94 |
180.36 ±48.64 |
0.098 |
87.37 ± 9.79 |
89.33 ± 9.04 |
96.00 |
0.637 |
HbA1c (%) |
5.76 ± 0.40 |
8.23 ± 1.76 |
7.61 ± 1.39 |
0.049* |
4.69 ± 0.37 |
4.75 ± 0.33 |
5.00 |
0.700 |
TG (mg/dL) |
146.67 ± 33.18 |
202.32 ± 34.07 |
165.73 ±36.31 |
0.140 |
164.31 ± 50.23 |
237.33 ± 88.06 |
199.00 |
0.026* |
TC (mg/dL) |
146.67 ± 33.18 |
202.32 ± 34.07 |
175.73 ±42.89 |
0.022* |
199.21 ± 40.24 |
220.83 ± 62.89 |
224.00 |
0.474 |
HDL (mg/dL) |
56.33 ± 16.92 |
49.08 ± 10.73 |
46.64 ± 9.54 |
0.398 |
47.24 ± 11.69 |
49.33 ± 10.94 |
35.00 |
0.525 |
LDL (mg/dL) |
185.33 ± 50.40 |
152.20 ± 48.20 |
134.36 ±59.00 |
0.428 |
144.34 ± 48.85 |
164.67 ± 52.34 |
231.00 |
0.181 |
BMI (kg/m2) |
32.77 ± 8.73 |
33.17 ± 9.11 |
34.08 ± 5.51 |
0.428 |
32.74 ± 3.28 |
32.06 ± 1.87 |
30.00 |
0.256 |
Visfatin (ng/L) |
88.20 ± 28.01 |
91.27 ± 37.16 |
107.18 ±37.16 |
0.263 |
178.44 ± 71.067 |
144.92 ± 32.76 |
110.50 |
0.028* |
SBP (mmHg) |
81.33 ± 33.79 |
117.12 ± 4.63 |
123.34 ± 2.78 |
0.298 |
121.00±7.72 |
120.00 ± 0.00 |
120.00 |
0.94 |
DBP (mmHg) |
56.66 ± 23.33 |
75.60 ± 3.05 |
78.18 ± 3.25 |
0.147 |
80.34 ± 1.83 |
80.00 ± 0.00 |
80.00 |
0.892 |
Insulin (μIU/mL) † |
6.16 ± 2.95 |
17.84 ± 3.00 |
33.65 ± 14.03 |
0.203 |
6.02 ± 3.21 |
6.16 ± 1.56 |
9.56 |
0.849 |
HOMA † |
1.72 ± 0.82 |
8.00 ± 1.32 |
14.38 ± 5.90 |
0.135 |
32.06 ± 1.87 |
1.38 ± 0.92 |
2.26 |
0.755 |
WHR |
1.40 ± 0.76 |
1.01 ± 0.27 |
1.18 ± 0.48 |
0.175 |
54.35 ± 33.74 |
69.45 ± 29.12 |
95.57 |
0.320 |
Fat mass (%) |
66.18 ± 29.17 |
47.14 ± 16.72 |
57.15 ±28.15 |
0.212 |
1.36 ± 0.63 |
1.61 ± 0.72 |
2.30 |
0.259 |
Serum visfatin level and its correlation with measured parameters
In non-diabetic obese women there were no significant correlation between parameters and serum visfatin level except for HDL (r = 0.378, p=0.018). Likewise in diabetic obese women, there were no significant correlation between serum visfatin level and indicated values except for FBS (r = 0.447, p = 0.004) and TG (r = 0.390, p = 0.014) as a moderate positive correlation and SBP (r= -0.520, P = 0.001) (Table 4).
Table 4: The correlation of visfatin level with measured parameters in diabetic and non-diabetic obese women.
parameters |
||||
|---|---|---|---|---|
Diabetic obese women |
Non-diabetic obese women |
|||
P value |
r value |
P value |
r value |
|
Age (year) |
0.787 |
0.045 |
0.122 |
0.263 |
BMI (kg/m2) |
0.753 |
0.053 |
0.555 |
0.102 |
FBS (mg/dL) |
0.004* |
0.447 |
0.440 |
0.133 |
HbA1c |
0.064 |
0.300 |
0.374 |
0.153 |
TG (mg/dL) |
0.014* |
0.390 |
0.053 |
-0.321 |
TC(mg/dL) |
0.385 |
0.143 |
0.480 |
-0.122 |
SBP (mmHg) |
0.001* |
-0.520 |
0.298 |
-0.171 |
DBP (mmHg) |
0.593 |
-0.092 |
0.267 |
0.100 |
HDL (mg/dL) |
0.16 |
0.525 |
0.018* |
0.378 |
LDL (mg/dL) |
0.119 |
-0.265 |
0.360 |
-0.151 |
Insulin (μIU/mL) † |
0.653 |
0.074 |
0.162 |
0.238 |
Fat mass (%) |
0.847 |
0.306 |
0.058 |
0.033 |
HOMA |
0.632 |
0.79 |
0.163 |
0.237 |
WHR |
0.066 |
0.298 |
0.835 |
0.038 |
Discussion
According to the previous reports, obesity is known as a key factor for developing T2DM and also insulin resistance. In addition association between obesity degree and its duration was found with incident of diabetes [54,55]. Hsiao et al. in a study indicated that rs9939609 variation in FTO gene has been significantly postulated to influence the obesity risk despite the inconsistency in its genetic effects on obesity [56]. Likewise Yajnik et al. demonstrated that association between variants of FTO gene and T2DM in Asian Indians was not completely mediated by BMI and adiposity. They concluded that BMI is a poor factor in measuring obesity in South Asian Indians [27]. Recently, certain studies on Japanese and Chinese populations failed to prove any association between FTO variants and T2DM, however, only in Japanese, just BMI was found to be in week association with FTO variants [20,21]. Asians in general are well known to have lower BMI comparing to Europeans and the relationship between risk of T2DM and obesity is sharper too [57]. In their study, Li et al. confirmed that FTO gene variations are in association (P<0.05) with enhanced risk of T2DM, in which, unlike Europeans, abolishment after adjustment for BMI is not observed in both East and South Asians [14]. Similar to discussed studies, in the present study, we demonstrated that in both diabetic and non-diabetic obese women FTO variant is not associated with BMI (P>0.05). The result was inconsistent with the results of Kara et al., who indicated that single nucleotide polymorphisms in FTO gene are in association with BMI [58]. Likewise, Quan et al. concluded that rs9939609 SNP is associated with increased risk of obesity in children and adolescents carrying homozygote genotypes [59], results which are in contrast with our findings.
These various outcomes may be related to different ethnic of the studied patients. Li et.al indicated significant small role of FTO gene on T2DM when adjusted with BMI in Asian than Europeans which is because of different adiposity phenotypes of these two populations [14]. Studies conducted by Li et al in 2008 and Fawad et al. in 2016 revealed no association between FTO rs9939609 SNP and obesity risk in Pakistani, Chinese, and Korean populations [20,23]. The frequency of A allele of investigated SNP in this study was 60.26% in diabetic and 10.81% in non-diabetic subjects, indicating its frequent incidence in diabetic ones. Overall, its frequency was 0.36, which is similar to certain populations like North Indian (0.31) and Pakistani (0.40) [23]. On the other hand, in Japanese and Chinese populations low frequency of this SNP has been observed [60]. Such discrepancy in results is likely due to substantial ethnic variation of the FTO rs9939609 SNP.
Furthermore, considering visfatin, it can be indicated that although exact biological mechanisms for visfatin mediated pathogenesis of T2DM is not known yet, based on its insulinmimetic influences visfatin can play important role as like as insulin in diabetic mice [44]. Chen et al. in 2005 conducted a study on visfatin and indicated that its elevated levels in T2DM might propose the visfatin signaling defect in certain tissues or any malfunction in biogenesis. They also reported no correlation between plasma visfatin level with BMI and any other parameters such as blood pressure, and lipid profile [44]. It has been reported that there is a positive correlation between obesity derived enhanced levels of visfatin and pancreatic cell damages and disruptions in insulin secretion [61]. Also, it was determined that plasma visfatin levels are elevated in obesity and various degrees of insulin resistance that indicates its special role in insulin resistance deficiency [62]. Consistently, our results suggest positive correlation between visfatin level and FBS in diabetic subjects whereas such relationship in non-diabetics was not observed. We demonstrated that visfatin concentrations in serum of diabetic patients are obviously enhanced in line with elevated levels of FBS. Similarly, Chen et al. and Wang et al. two independent studies in 2006 suggested significant raised levels of visfatin in plasma of T2DM patients [44,63]. Moreover, Wang et al. indicated significant correlation between plasma visftain level and lipid profile. Considering this study, visfatin was in correlation with high HDL and low TG [63]. In contrast, our results reveal no correlation between such lipid profiles and visfatin levels. In line with previous studies held by De Luis et al. and Jian et al. [64,65], this study proposes positive correlation between visfatin level with TG levels in diabetic obese women. In this study we clarified that visfatin level is significantly lower in diabetic obese women than non-diabetic obese ones, the results which are overall similar to those of Jian et al. [65]. According to Sethi et al., there are expected correlations between vsifatin level and indexes such as BMI and WHR as visfatin is secreted from adipose tissue [66], however, in the present study, such correlations were not observed.
Putting the discussion in a wider context and considering probable association between re9939609 SNP and visfatin level, Lopez et al. in 2008 investigated the association of cord visfatin with this SNP in 234 neonatal, which was in line with the result of previous studies indicating the potential interaction between FTO and visfatin gene expression. They declared the possible role of FTO in regulation of visfatin expression in adipose tissue through DNA demethylation [67]. According to these various evidences, it is demonstrated that frequency of FTO rs9939609 is different between various races of people but FTO was remained as a main locus which showed association with obesity and T2DM.
Limitations of the study
The report of study was a result of small sample size of diabetic obese women. It is suggested that the study could be perform on larger sample size population. In addition, due to limited number of subjects with homozygote variations, testTukey was not applied.
Conclusion
Association of FTO gene variants especially rs3399609 with risk of obesity and T2DM was determined in various populations. Our results also showed this association with high frequency of rs3399609 in diabetic obese women. In addition, researches are focused on visfatin link with obesity and T2DM which was secreted from adipose tissue. This study confirmed this link and determined that the mean of visfatin level in diabetic women who harbored rs3399609 was high than wild type genotype (TT). Finally, it is propose that visfatin level could be used as a significant indicator in diabetic obese women.
Declarations
Acknowledgements: This research was granted by Infectious and Tropical Disease research center, Tabriz University of Medical Sciences, Iran (No: 19864/5). Authors are grateful to Department of Clinical Biochemistry and Laboratory Medicine, Faculty of Medicine, Tabriz University of Medical Sciences, Iran, for their support.
Author’s contribution: Writing - original draft preparation: Davoud Jafari-gharabaghlou. Editing: Davoud Jafari-gharabaghlou and Nosratollah Zarghami. Conceptualization, Supervision: Nosratollah Zarghami.
Consent to publication: Informed consent was obtained from all individual participants included in the study.
Availability of supporting data: The data that support the findings of this study are available from the corresponding author, upon reasonable request.
Competing Interests: No potential competing interest was reported by the authors.
References
- Hossain P, Kawar B, El Nahas M. Obesity and diabetes in the developing world—a growing challenge. New England Journal of Medicine. 2007; 356: 213-215.
- Ogden CL, Yanovski SZ, Carroll MD, Flegal KM. The epidemiology of obesity. Gastroenterology. 2007; 132: 2087-2102.
- Esfahlan RJ, Zarghami N, Esfahlan AJ, Mollazadeh M, Nejati K, Nasiri M. The Possible Impact of Obesity on Androgen, Progesterone and Estrogen Receptors (ERα and ERβ) Gene Expression in Breast Cancer Patients. Breast Cancer : Basic and Clinical Research. 2011; 5: 227-237.
- Andolfi C, Fisichella PM. Epidemiology of obesity and associated comorbidities. Journal of Laparoendoscopic & Advanced Surgical Techniques. 2018; 28: 919-924.
- Ben Halima M, Kallel A, Baara A, Ben Wafi S, Sanhagi H, Slimane H, et al. The rs9939609 polymorphism in the fat mass and obesity associated (FTO) gene is associated with obesity in Tunisian population. Biomarkers. 2018; 23: 787-792.
- Gerken T, Girard CA, Tung Y-CL, Webby CJ, Saudek V, Hewitson KS, et al. The obesity-associated FTO gene encodes a 2-oxoglutarate-dependent nucleic acid demethylase. Science. 2007; 318: 1469-1472.
- Peters T, Ausmeier K, Dildrop R, Rüther U. The mouse Fused toes (Ft) mutation is the result of a 1.6-Mb deletion including the entire Iroquois B gene cluster. Mammalian genome. 2002; 13: 186- 188.
- Frayling TM, Timpson NJ, Weedon MN, Zeggini E, Freathy RM, Lindgren CM, et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science (New York, NY). 2007; 316: 889-894.
- Jalili V, Mokhtari Z, Rastgoo S, Hajipour A, Bourbour F, Gholamalizadeh M, et al. The association between FTO rs9939609 polymorphism and serum lipid profile in adult women. Diabetology & Metabolic Syndrome. 2021; 13: 1-6.
- Hinney A, Nguyen TT, Scherag A, Friedel S, Brönner G, Müller TD, et al. Genome wide association (GWA) study for early onset extreme obesity supports the role of fat mass and obesity associated gene (FTO) variants. PloS one. 2007; 2: e1361.
- Scuteri A, Sanna S, Chen W-M, Uda M, Albai G, Strait J, et al. Genome-wide association scan shows genetic variants in the FTO gene are associated with obesity-related traits. PLoS Genet. 2007; 3: e115.
- Yusof H, Osmera AN, Hatta FHM. A Review on the Association of Fat Mass Obesity Associated (FTO) Gene Marker Rs9939609 with Obesity and Body Mass Index (BMI). Buletin Sains Kesihatan. 2021; 5: 11-20.
- Speliotes EK, Willer CJ, Berndt SI, Monda KL, Thorleifsson G, Jackson AU, et al. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nature genetics. 2010; 42: 937-948.
- Li H, Kilpeläinen TO, Liu C, Zhu J, Liu Y, Hu C, et al. Association of genetic variation in FTO with risk of obesity and type 2 diabetes with data from 96,551 East and South Asians. Diabetologia. 2012; 55: 981-995.
- Zeggini E, Weedon MN, Lindgren CM, Frayling TM, Elliott KS, Lango H, et al. Replication of genome-wide association signals in UK samples reveals risk loci for type 2 diabetes. Science. 2007; 316: 1336-1341.
- Andreasen CH, Stender-Petersen KL, Mogensen MS, Torekov SS, Wegner L, Andersen G, et al. Low physical activity accentuates the effect of the FTO rs9939609 polymorphism on body fat accumulation. Diabetes. 2008; 57: 95-101.
- Peeters A, Beckers S, Verrijken A, Roevens P, Peeters P, Van Gaal L, et al. Variants in the FTO gene are associated with common obesity in the Belgian population. Molecular genetics and metabolism. 2008; 93: 481-484.
- Price RA, Li W-D, Zhao H. FTO gene SNPs associated with extreme obesity in cases, controls and extremely discordant sister pairs. BMC Medical Genetics. 2008; 9: 1.
- Legry V, Cottel D, Ferrières J, Arveiler D, Andrieux N, Bingham A, et al. Effect of an FTO polymorphism on fat mass, obesity, and type 2 diabetes mellitus in the French MONICA Study. Metabolism. 2009; 58: 971-975.
- Li H, Wu Y, Loos RJ, Hu FB, Liu Y, Wang J, et al. Variants in the fat mass–and obesity-associated (FTO) gene are not associated with obesity in a Chinese Han population. Diabetes. 2008; 57: 264-268.
- Hotta K, Nakata Y, Matsuo T, Kamohara S, Kotani K, Komatsu R, et al. Variations in the FTO gene are associated with severe obesity in the Japanese. Journal of human genetics. 2008; 53: 546-553.
- Ng MC, Park KS, Oh B, Tam CH, Cho YM, Shin HD, et al. Implication of genetic variants near TCF7L2, SLC30A8, HHEX, CDKAL1, CDKN2A/B, IGF2BP2, and FTO in type 2 diabetes and obesity in 6,719 Asians. Diabetes. 2008; 57: 2226-2233.
- Fawwad A, Siddiqui IA, Basit A, Zeeshan NF, Shahid SM, Nawab SN, et al. Common variant within the FTO gene, rs9939609, obesity and type 2 diabetes in population of Karachi, Pakistan. Diabetes & Metabolic Syndrome: Clinical Research & Reviews. 2016; 10: 43-47.
- Cirillo E, Kutmon M, Gonzalez Hernandez M, Hooimeijer T, Adriaens ME, Eijssen LM, et al. From SNPs to pathways: Biological interpretation of type 2 diabetes (T2DM) genome wide association study (GWAS) results. PloS one. 2018; 13: e0193515.
- Hertel JK, Johansson S, Sonestedt E, Jonsson A, Lie RT, Platou CG, et al. FTO, Type 2 Diabetes, and Weight Gain Throughout Adult Life A Meta-Analysis of 41,504 Subjects From the Scandinavian HUNT, MDC, and MPP Studies. Diabetes. 2011; 60: 1637-1644.
- Liu Y, Liu Z, Song Y, Zhou D, Zhang D, Zhao T, et al. Meta‐analysis added power to identify variants in FTO associated with type 2 diabetes and obesity in the Asian population. Obesity. 2010; 18: 1619-1624.
- Yajnik CS, Janipalli CS, Bhaskar S, Kulkarni SR, Freathy RM, Prakash S, et al. FTO gene variants are strongly associated with type 2 diabetes in South Asian Indians. Diabetologia. 2009; 52: 247-252.
- Li X, Song F, Jiang H, Zhang M, Lin J, Bao W, et al. A genetic variation in the fat mass‐and obesity‐associated gene is associated with obesity and newly diagnosed type 2 diabetes in a Chinese population. Diabetes/metabolism research and reviews. 2010; 26: 128-132.
- Sanghera DK, Ortega L, Han S, Singh J, Ralhan SK, Wander GS, et al. Impact of nine common type 2 diabetes risk polymorphisms in Asian Indian Sikhs: PPARG2 (Pro12Ala), IGF2BP2, TCF7L2 and FTO variants confer a significant risk. BMC Medical Genetics. 2008; 9: 1.
- Takeuchi F, Yamamoto K, Katsuya T, Nabika T, Sugiyama T, Fujioka A, et al. Association of genetic variants for susceptibility to obesity with type 2 diabetes in Japanese individuals. Diabetologia. 2011; 54: 1350-1359.
- Bo X, Jie M. FTO polymorphisms are associated with obesity but not with diabetes in East Asian populations: a meta-analysis. Biomedical and Environmental Sciences. 2009; 22: 449-457.
- Ramya K, Radha V, Ghosh S, Majumder PP, Mohan V. Genetic variations in the FTO gene are associated with type 2 diabetes and obesity in south Indians (CURES-79). Diabetes technology & therapeutics. 2011; 13: 33-42.
- Omori S, Tanaka Y, Takahashi A, Hirose H, Kashiwagi A, Kaku K, et al. Association of CDKAL1, IGF2BP2, CDKN2A/B, HHEX, SLC30A8, and KCNJ11 with susceptibility to type 2 diabetes in a Japanese population. Diabetes. 2008; 57: 791-795.
- Shu XO, Long J, Cai Q, Qi L, Xiang Y-B, Cho YS, et al. Identification of new genetic risk variants for type 2 diabetes. PLoS Genet. 2010; 6: e1001127.
- Mohammadzadeh G, Zarghami N. Serum leptin level is reduced in non-obese subjects with type 2 diabetes. Int J Endocrinol Metab. 2013; 11: 3-10.
- Mohammadzadeh G, Zarghami N. Associations between singlenucleotide polymorphisms of the adiponectin gene, serum adiponectin levels and increased risk of type 2 diabetes mellitus in Iranian obese individuals. Scandinavian journal of clinical and laboratory investigation. 2009; 69: 764-771.
- Mofarrah M, Ziaee S, Pilehvar-Soltanahmadi Y, Zarghami F, Boroumand M, Zarghami N. Association of KALRN, ADIPOQ, and FTO gene polymorphism in type 2 diabetic patients with coronary artery disease: possible predisposing markers. Coron Artery Dis. 2016; 27: 490-496.
- Zorena K. Obesity and Type 2 Diabetes Mellitus: Adipocytokines as Markers of Insulin Resistance. Frontiers in Clinical Drug Research: Diabetes and Obesity. 2016; 3: 195.
- Kang YE, Kim JM, Joung KH, Lee JH, You BR, Choi MJ, et al. The Roles of Adipokines, Proinflammatory Cytokines, and Adipose Tissue Macrophages in Obesity-Associated Insulin Resistance in Modest Obesity and Early Metabolic Dysfunction. PLoS ONE. 2016; 11: e0154003.
- Mohammadzadeh G, Zarghami N. Hypoadiponectinemia in obese subjects with type II diabetes: A close association with central obesity indices. Journal of research in medical sciences : the official journal of Isfahan University of Medical Sciences. 2011; 16: 713-723.
- Conde J, Scotece M, Gomez R, Lopez V, Gomez-Reino JJ, Lago F, et al. Adipokines: biofactors from white adipose tissue. A complex hub among inflammation, metabolism, and immunity. BioFactors (Oxford, England). 2011; 37: 413-420.
- Jafari-Gharabaghlou D, Vaghari-Tabari M, Oghbaei H, Lotz L, Zarezadeh R, Rezaei YR, et al. Role of adipokines in embryo implantation. Endocrine connections. 2021; 10: R267-R278.
- Akbarian N, Zarghami N, Mota A, Abediazar S, Abroon S, Mihanfar A, et al. Correlation between circulating visfatin and nitric oxide metabolites levels in patients with diabetic nephropathy. Iranian journal of kidney diseases. 2018; 12: 163.
- Chen MP, Chung FM, Chang DM, Tsai JCR, Huang HF, Shin SJ, et al. Elevated plasma level of visfatin/pre-B cell colony-enhancing factor in patients with type 2 diabetes mellitus. The Journal of Clinical Endocrinology & Metabolism. 2006; 91: 295-299.
- Ghaemmaghami S, Hedayati M, Mohaddes SM, Mohammadi MG, Rahmati M, Zarghami N. Visfatin proliferative effect on HCT116 colorectal cancer cell line. Advances in Environmental Biology. 2014: 55-61.
- Mohammadi M, Zarghami N, Hedayati M, Ghaemmaghami S, Yamchi RM, Mohaddes M. Visfatin effects on telomerase gene expression in AGS gastric cancer cell line. Indian journal of cancer. 2015; 52: 32-35.
- Mohammadi M, Zarghami N, Hedayati M, Ghaemmaghami S. Synergistic Effects of Resistin and Visfatin as Adipocyte Derived Hormones on Telomerase Gene Expression in AGS Gastric Cancer Cell Line. Acta medica Iranica. 2017; 55: 621-627.
- Organization WH. Classification of diabetes mellitus. 2019.
- Dairawan M, Shetty PJ. The evolution of DNA extraction methods. America Journal of Biomedical Science and Research. 2020; 8: 39-46.
- Majdi MA, Mohammadzadeh NA, Lotfi H, Mahmoudi R, Alipour FG, Shool F, et al. Correlation of Resistin Serum Level with Fat Mass and Obesity-Associated Gene (FTO) rs9939609 Polymorphism in Obese Women with Type 2 Diabetes. Diabetes & metabolic syndrome 2017.
- https://blast.ncbi.nlm.nih.gov/Blast.cgi BLASTAf.
- Friedewald WT, Levy RI, Fredrickson DS. Estimation of the concentration of low-density lipoprotein cholesterol in plasma, without use of the preparative ultracentrifuge. Clinical chemistry. 1972; 18: 499-502.
- Jackson AS, Stanforth PR, Gagnon J, Rankinen T, Leon AS, Rao DC, et al. The effect of sex, age and race on estimating percentage body fat from body mass index: The Heritage Family Study. International journal of obesity and related metabolic disorders: journal of the International Association for the Study of Obesity. 2002; 26: 789-796.
- Steppan CM, Lazar MA. Resistin and obesity-associated insulin resistance. Trends in endocrinology & Metabolism. 2002; 13: 18- 23.
- Day C, Bailey CJ. Obesity in the pathogenesis of type 2 diabetes. The British Journal of Diabetes & Vascular Disease. 2011; 11: 55- 61.
- Hsiao TJ, Lin E. Association of a common rs9939609 variant in the fat mass and obesity-associated (FTO) gene with obesity and metabolic phenotypes in a Taiwanese population: a replication study. Journal of genetics. 2016; 95: 595-601.
- Ramachandran A, Snehalatha C, Viswanathan V, Viswanathan M, Haffner S. Risk of noninsulin dependent diabetes mellitus conferred by obesity and central adiposity in different ethnic groups: a comparative analysis between Asian Indians, Mexican Americans and Whites. Diabetes research and clinical practice. 1997; 36: 121-125.
- Karra E, O’Daly OG, Choudhury AI, Yousseif A, Millership S, Neary MT, et al. A link between FTO, ghrelin, and impaired brain food-cue responsivity. The Journal of clinical investigation 2013; 123: 3539-3551.
- Quan L, Wang H, Tian Y, Mu X, Zhang Y, Tao K. Association of fat-mass and obesity-associated gene FTO rs9939609 polymorphism with the risk of obesity among children and adolescents: a meta-analysis. Eur Rev Med Pharmacol Sci. 2015; 19: 614-623.
- Prakash J, Mittal B, Srivastava A, Awasthi S, Srivastava N. Association of FTO rs9939609 SNP with Obesity and Obesity-Associated Phenotypes in a North Indian Population. Oman medical journal. 2016; 31: 99.
- López-Bermejo A, Chico-Julià B, Fernàndez-Balsells M, Recasens M, Esteve E, Casamitjana R, et al. Serum visfatin increases with progressive β-cell deterioration. Diabetes. 2006; 55: 2871-2875.
- Ghanbari-Niaki A, Saghebjoo M, Soltani R, Kirwan JP. Plasma visfatin is increased after high-intensity exercise. Annals of Nutrition and Metabolism. 2010; 57: 3-8.
- Wang T, Zhang X, Bheda P, Revollo JR, Imai S-i, Wolberger C. Structure of Nampt/PBEF/visfatin, a mammalian NAD+ biosynthetic enzyme. Nature structural & molecular biology.2006; 13.
- de Luis DA, Sagrado MG, Aller R, Conde R, Izaola O. Circulating visfatin in obese non-diabetic patients in relation to cardiovascular risk factors, insulin resistance, and adipocytokines: A contradictory piece of the puzzle. Nutrition. 2010; 26: 1130-1133.
- Jian WX, Luo TH, Gu YY, Zhang HL, Zheng S, Dai M, et al. The visfatin gene is associated with glucose and lipid metabolism in a Chinese population. Diabetic Medicine. 2006; 23: 967-973.
- Sethi JK, Vidal-Puig A. Visfatin: the missing link between intraabdominal obesity and diabetes? Trends in molecular medicine. 2005; 11: 344-347.
- López-Bermejo A, Petry CJ, Díaz M, Sebastiani G, de Zegher F, Dunger DB, et al. The association between the FTO gene and fat mass in humans develops by the postnatal age of two weeks. The Journal of Clinical Endocrinology & Metabolism. 2008; 93: 1501-1505.